What Is Stellaris RNA FISH?
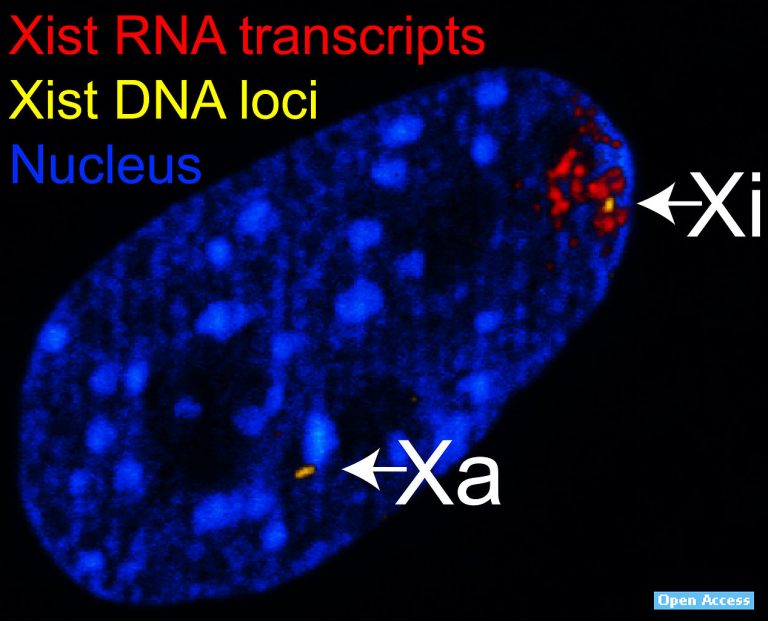 Stellaris RNA FISH: Detection, Localization, and Quantification of RNA at the Sub- Cellular Level
Stellaris RNA FISH: Detection, Localization, and Quantification of RNA at the Sub- Cellular LevelStellaris® RNA FISH (fluorescence in situ hybridization) is a RNA visualization method that allows simultaneous detection, localization, and quantification of individual RNA molecules at the sub-cellular level in fixed samples using wide-field or confocal fluorescence microscopy. This novel RNA FISH technology represents a fast and easy to use method to achieve conclusive results through compelling images of RNA expressions. A set of Stellaris RNA FISH Probes comprises multiple oligonucleotides with different sequences each with a fluorescent label that collectively bind along the same target transcript to produce a punctate signal.
The Stellaris RNA FISH technology follows a simple protocol, does not use exotic reagents, is inexpensive and platform-independent. It also offers same-day results, and is versatile both in terms of sample type and application. Stellaris RNA FISH probes may even be labeled with several different dyes, to enable simultaneous multiplex detection of different RNA targets. Finally, scientists can address the stochastic nature of gene expression and visualize RNA through direct detection without isolation, purification, and amplification.
PROBES
1. Custom Probe Sets
Design your own Custom RNA FISH Probe Set to detect your RNA of interest. You can design up to 48 individually fluorescently labeled oligonucleotides to bind along and visualize your target RNA.
2. ShipReady Probe Sets
ShipReady control probe sets for Stellaris® RNA FISH is significantly convenient, ready to use positive controls for mouse or human reference and long non-coding RNA targets. Both qualitative (localization) and quantitative (transcript counting) controls are available in the most commonly used model organisms including Human, Mouse, C. elegans, Drosophila, and Yeast. All Stellaris ShipReady products have been verified on cultured cells or whole mount tissues to give you confidence in your studies.
3. DesignReady Probe Sets
DesignReady Stellaris® RNA FISH Probe Sets take the guesswork out of designing your own custom assay. These professionally designed sets go through rigorous pre- and post-design bioinformatic analysis to maximize specificity and most are available in the following dyes: CAL Fluor® Red 590, Quasar® 570, CAL Fluor Red 610, Quasar 670, CAL Fluor Red 635, CAL Fluor Orange 560, and FAM.
4. Chromatin Isolation Via RNA Precipitation (ChIRP)
ChIRP is a rapid technique to discover RNA-associated DNA sequences and map genomic binding sites of chromatin associated long noncoding RNAs (lncRNAs) with high sensitivity and low background. This method allows you to map which chromatin regions come together for coordinated expression, determine which genes are expressed together, and learn the role of lncRNA in chromatin regulation. Target lncRNAs are affinity captured using antisense-oligos, designed by our ChIRP probe designer, and the lncRNA-associated DNA chromatin is sequenced to create a sequencing library. With the DNA sequence data, it is possible to generate a genomic binding site map at a resolution of several hundred bases.
5. ChIRP Probe Designer
ChIRP oligo sets can be designed at LGC Biosearch’s dedicated web-based designer and ordered directly. The oligos are delivered in plates so that they may be combined into suitable sets. We recommend buffers and other reagents for the complete ChIRP method provided by EMD/Millipore. LGC Biosearch Technologies and EMD/Millipore are exclusively licensed to provide ChIRP probe sets.
6. RNA FISH Buffers
Stellaris® RNA FISH Buffers contain proprietary additives to enhance detection with Stellaris RNA FISH probe sets, particularly for applications that typically exhibit more pronounced background fluorescence.
Stellaris RNA FISH Buffers outperform Conventional RNA FISH buffers
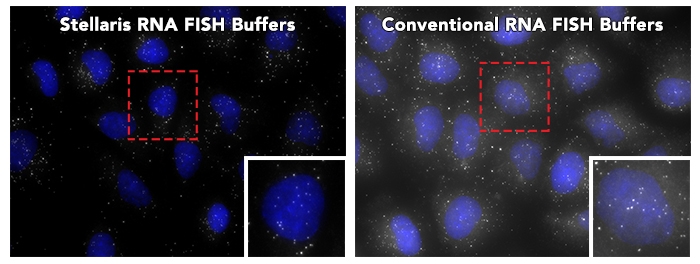
A direct comparison showing background reduction by using Stellaris RNA FISH Buffers (Cat#: SMF-HB1-10, SMF-WA1-60, SMF-WB1-20). Following the adherent cell protocol, formalin fixed human cells were probed with Human POLR2A (Cat# VSMF-2293-5). Left Panel: RNA FISH performed with Stellaris RNA FISH Buffers. Right Panel: RNA FISH performed with Conventional RNA FISH buffer recipes. Note: Results may vary according to original probe set performance and sample type.
Stellaris RNA FISH Protocol
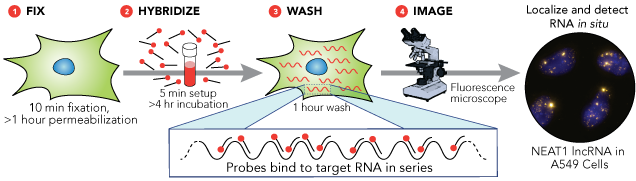
The Stellaris RNA FISH protocol is comparatively simple, and consists of four steps, as shown in the image below. No exotic reagents are required, and the entire process can be completed in less than a day.
Step 1: Prepare Sample
The sample is grown on or adhered, or sectioned onto a #1 coverglass and permeabilized with 70% ethanol. Slight variations in sample preparation between different organisms and sample types are covered in the online protocols. No protease is required.
Step 2: Hybridize Probes
For most samples; hybridization can be completed in 4 hours at +37 °C in a generic laboratory incubator.
Step 3: Wash Sample
After hybridization, wash buffers with short incubation periods are used to remove excess probes. The total time for this step is 1 – 1.5 hours.
Step 4: Image Sample
At this point, the sample can be imaged using a standard fluorescence microscope. We recommend using a widefield fluorescence microscope though confocal microscopy can also be used with Stellaris RNA FISH probes.
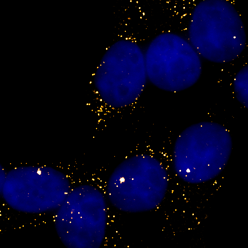
Sensitivity & Specificity
The large number of probes in a Stellaris RNA FISH assay ensures both high sensitivity and specificity. The inherent redundancies of this direct detection method minimizes the possibility of false negative and false positive signals. Positive signal identified from the combined localized fluorescence of multiple probes. Off-target binding of single probes generates only weak and diffuse fluorescence, well below the threshold for detection of the specifically-targeted mRNAs. Furthermore, Stellaris RNA FISH probes can bind to partially degraded target mRNAs making them well-suited for formalin-fixed, paraffin-embedded (FFPE) tissue samples.
Multiplexing
Stellaris RNA FISH probes are available with a wide variety of fluorophores, including Biosearch Technologies’ CAL Fluor® and Quasar® dyes and can be used in a multiplex assay. The dye choices in a multiplex assay are largely determined by the filters available to a researcher, but it is certainly possible to design an assay including a DAPI nuclear stain and three Stellaris RNA FISH probe sets labeled with reporter dyes with minimal spectral overlap.
Multiplex assays can be used to identify a correlation in expression levels between multiple genes, run a control probe set of known expression with a target simultaneously, or simply save time over running the assays one at a time. For human cells, we offer inventoried probe sets suitable for use as positive controls. These foundation gene probe sets are labeled with Quasar® 570 dye.
Stellaris RNA FISH can also be combined with existing technologies such as qPCR, DNA FISH, IHC (immunohistochemistry), and western blotting to provide complementary information.

