Pooled CRISPR guide RNA libraries, or gRNA libraries, are ideal for high-throughput screening of important molecular targets. These libraries leverage the efficiency and specificity of the CRISPR gene editing technology to either knock-out gene expression or transcriptionally activate genes in the genome.
We offers a variety of gRNA library options, all of which contain sequences pre-validated, including (1) GeCKO libraries for genome-wide knockout of human and mouse genes, (2) CRISPR Synergistic Activating Mediator (SAM) libraries for transcriptional activation of every gene in the human and mouse genome, and (3) Pathway-focused gRNA libraries that genes with a specific function.
How are CRISPR libraries used?
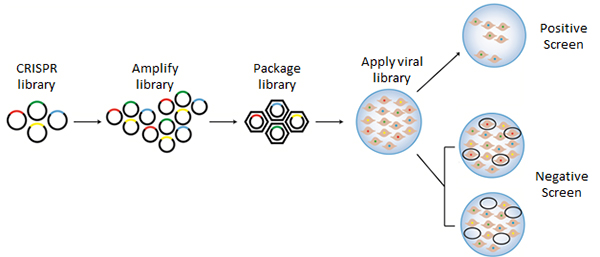
Screening using the CRISPR technology is particularly advantageous because of its simplicity, specificity and versatility. The genome-wide GeCKO and SAM libraries were designed to target every gene in the mouse or human genes and knock-out or transcriptionally activate each gene, respectively. Pathway-focused gRNA libraries were designed from targets identified through the Drug Gene Interaction Database, providing application-focused gRNA libraries for more simplified screening. Libraries are amplified, packaged and transduced into host cells to generate mutant cell lines (Figure 1). The mutants can then be screened under specific conditions, such as by positive or negative selection, to identify important genes in a pathway.
The genome-scale CRISPR knock-out (GeCKO) v2 libraries enable rapid screening for loss-of-function mutations that produce a desired phenotype in human or mouse cells. We are pleased to offer all eight GeCKO v2 CRISPR libraries. GeCKO libraries are mixed pools of guide RNA targeting each gene and miRNA within the mouse or human genome; the improved GeCKO v2.0 libraries also contain non-targeting gRNA to serve as controls and utilize improved lentiviral vectors for high-titer lentivirus production and highly efficient transduction of primary cells. GeCKO libraries have been used in primary mouse and human cells, stem cells, cancer cells, and stable cell lines. GeCKO library screens allows you to identify genes that are essential for cell viability under different conditions, as well as genes whose loss enables drug resistance in cancer cells. For complete details of GeCKOv2 library design & construction
GeCKO v2 libraries are available as two half-libraries, A and B. Each library contains 3gRNAs per gene. Both libraries contain 1,000 non-targeting control sgRNAs. The A library contains gRNAs targeting miRNAs (4 sgRNAs per miRNA) as well. The A and B libraries can be purchased and delivered together to ensure complete representation; however, as more cells are required for both libraries, screening with library A is suggested.
GeCKO Library Deliverables
(a) Pooled library (Transfection Grade) in 25 µg aliquots. The 100 µg and 200 µg library are delivered as 4 x 25 µg or 8 x 25 µg, respectively. The libraries are delivered with a technical guide, MSDS and COA.
(b) Mouse GeCKO Library A (dual vector): 100 µg pooled library (Research Grade)
(c) Scale up to 900 µg total, in increments of 100 µg; Transfection Grade available instead of Research Grade
(d) Final project report, including ABI files from Sanger sequencing for small-scale tests and statistical summary from next-generation sequencing validation. Complete raw NGS data available upon request
How to use GeCKO libraries
GeCKO libraries are a mixed pool of CRISPR guide RNAs that target every gene and miRNA in the genome. Each gRNA is cloned into a lentiviral vector optimized to produce high-titer virus for efficient lentiviral transduction of primary cells or cultured cell lines.
CRISPR transcriptional activation SAM library
SAM libraries induce robust transcriptional activation for genome-wide gain-of-function screening
The SAM library enables gain-of-function screening in human cells by inducing robust, specific transcriptional activation of each coding region in the genome in a single screen. CRISPR/Cas9 Synergistic Activation Mediator (SAM) is a protein complex engineered to enable robust transcriptional activation of endogenous genes. SAM takes advantage of the specificity and ease of reprogramming of Cas9 nucleases, which are targeted to a specific locus in the genome by guide RNA. The SAM complex includes three transcription activation domains VP64, P65, and HSF1 which synergistically enhance transcription when targeted to any site within 200bp upstream of a transcription start site. SAM uses a nucleolytically inactive Cas9 to ensure that no strand breaks are introduced into endogenous genome.
The human genome-wide SAM library activates all known coding isoforms in the RefSeq database (23,430 isoforms) for gain-of-function screening. SAM libraries have been used in primary mouse and human cells, stem cells, cancer cells, and stable cell lines. SAM library screens allow you to identify genes that are essential for cell viability under different conditions, as well as genes whose loss enables drug resistance in cancer cells.
SAM Library Deliverables
All SAM libraries are Transfection Grade and in-stock for immediate delivery. Additional quantity options are available, but may require additional turnaround time. Email us for more information on additional library options.

